By Keisha Rose Harrison, MS (PhD Candidate at Oregon State University)
Kombucha, a funky fermented tea, is a time immemorial beverage that has made appearances across the globe in various forms for eons. This beverage of unknown origin is shrouded in many mysteries. The potential health benefits, fermentation kinetics, and alcohol production are a few aspects that continue to baffle kombucha lovers and producers alike. Meanwhile, one area where fermentation researchers are making ground is in defining the microbiology of the Kombucha SCOBY.
In comparison to other fermented beverages, i.e. wine and beer, the amount of research done on the Kombucha SCOBY is sparse. It was known in even the earliest studies, from back in the 1960s that the starter colony was made up of both yeast and bacteria. However, there was little known about which organisms drove the fermentation, contributed to flavor, and fought off spoilage. Until recently, scientists relied upon culture-based methods (isolating microbes on media plates) to identify yeast and bacteria organisms. This technique, although cost-effective and accessible, is prone to error. Despite its limitations, scientists (Teoh et al, 2004; Liu et al, 1996), were able to come to consensus about the role of acetic acid bacteria (AAB) and saccharolytic (“sugar eating”) yeast.
By the early 2000s, we understood that AAB converted ethanol to acetic acid and built the SCOBY. Acetobacter and Gluconoacterobacter, genera of AAB, were claimed as staples and have since been renamed to fit under the genus Komagataeibacter. There was less agreement about which yeast cells eat cane sugar to produce simple sugar hexoses for the bacteria. There are accounts of Zygosaccharomyces, Schizosaccharomyces, Brettanomyces, and Candida. However, Teoh (2004) warns us that identifying organisms based upon isolation methods is generally unsuccessful.
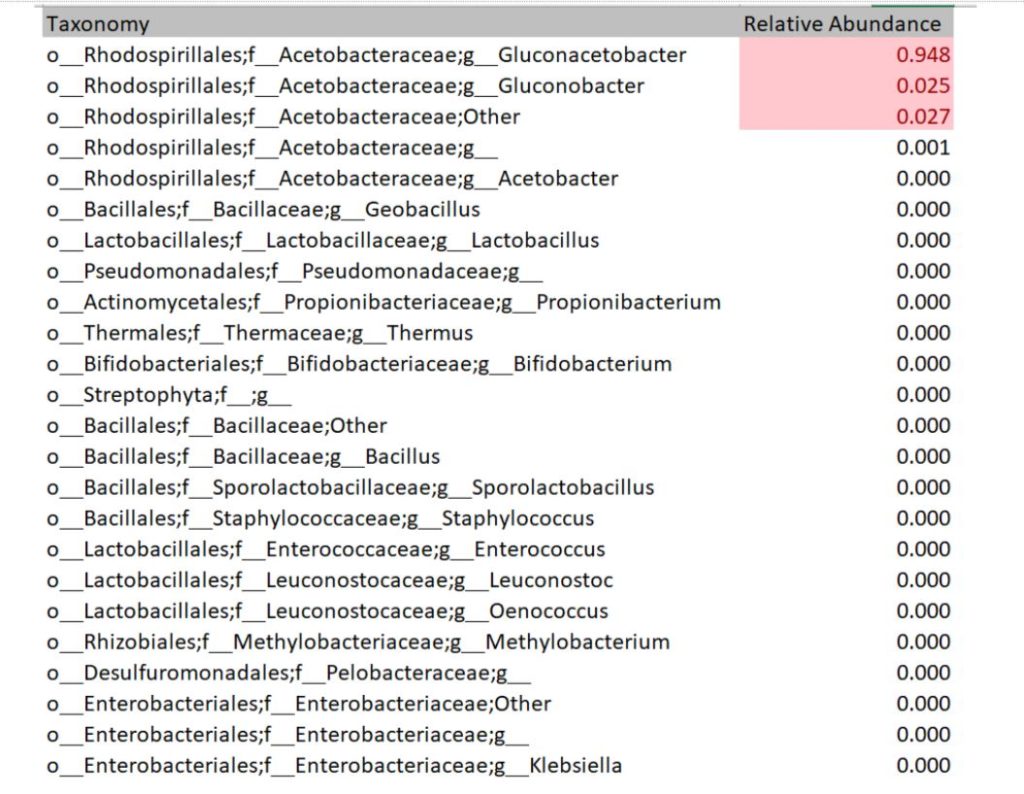
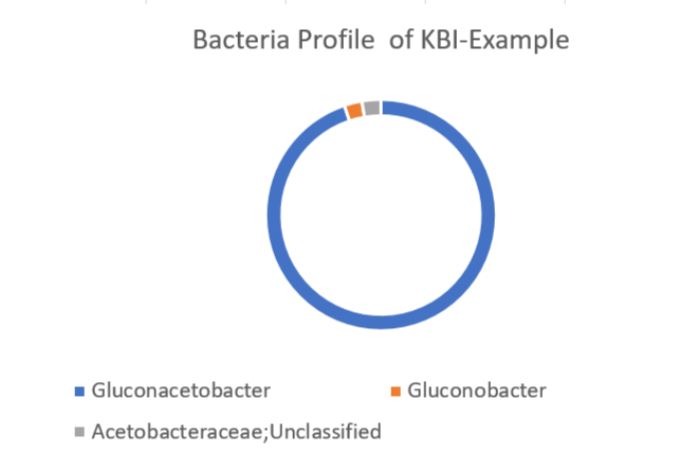
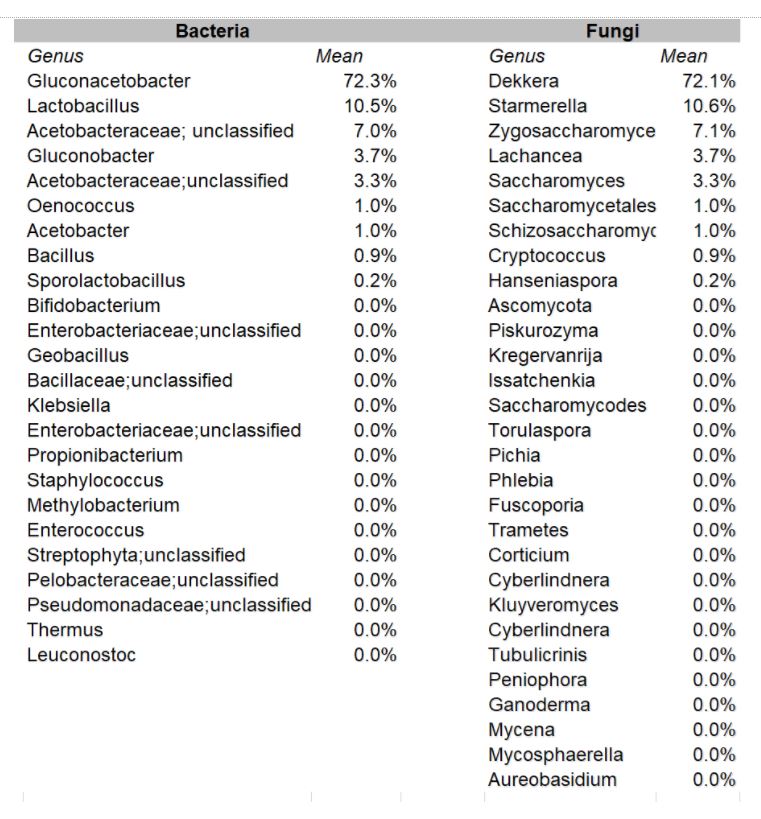
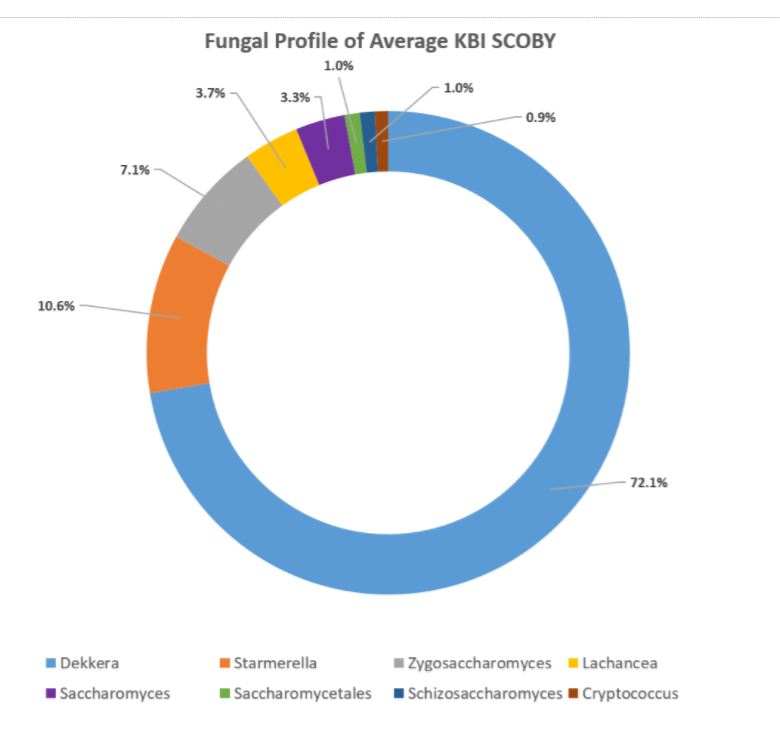
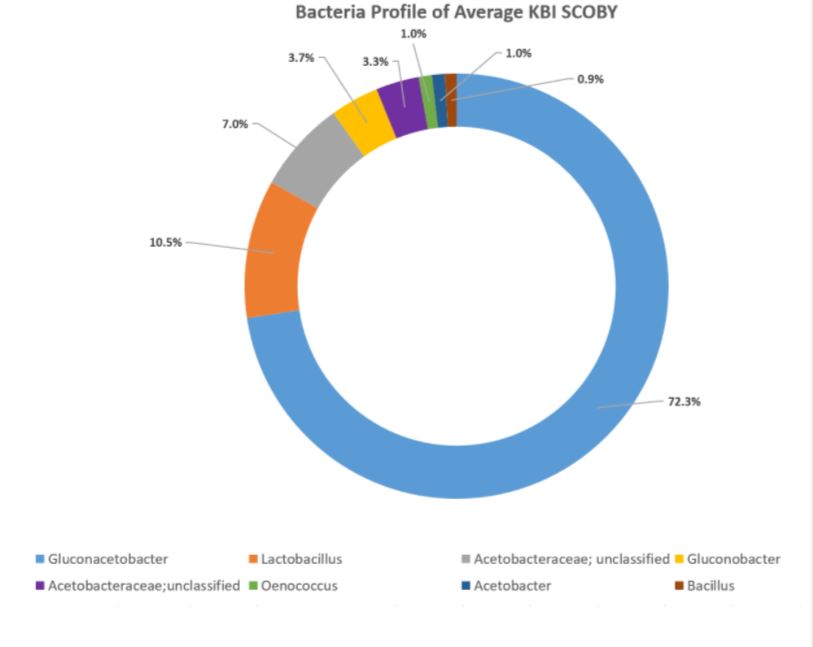
 The DNA sequencing boom changed the name of the game! With the advent of affordable and reliable genetic sequencing, high-resolution taxonomic identification became a thing of reality. First scientists used sequencing technology to characterize isolated organism as a more robust culture dependent approach. However, with next generation sequencing (NGS), we could finally for the first time identify both culturable and nonculturable organisms. With NGS technology, scientists confirmed Komagatebacter as the dominant Kombucha bacteria (Marsh, 2014; Chakravorty, 2016) revealed that lactic acid and thermophilic bacteria that are likely dependent upon Kombucha preparation and region. (See table below for comprehensive list of evaluated microorganisms) These studies finally give us an in-depth analysis of kombucha microflora.
The DNA sequencing boom changed the name of the game! With the advent of affordable and reliable genetic sequencing, high-resolution taxonomic identification became a thing of reality. First scientists used sequencing technology to characterize isolated organism as a more robust culture dependent approach. However, with next generation sequencing (NGS), we could finally for the first time identify both culturable and nonculturable organisms. With NGS technology, scientists confirmed Komagatebacter as the dominant Kombucha bacteria (Marsh, 2014; Chakravorty, 2016) revealed that lactic acid and thermophilic bacteria that are likely dependent upon Kombucha preparation and region. (See table below for comprehensive list of evaluated microorganisms) These studies finally give us an in-depth analysis of kombucha microflora.
What does this mean to brewers? The recent application of advanced sequencing proved that we can apply high-fidelity identification tools to kombucha. With the precedent already set, we can now focus on designing experiments that address the larger questions. For instance, “what is common to commercial SCOBY?” and “which organisms are found in kombucha with high organic acid production?” By sequencing a mass number of SCOBY from around the globe, we will have the potential to link microbes with the chemical composition of finished kombucha products. Additionally, we will get closer to curating reproducibility and troubleshooting problematic batches.
How you can get involved? KBI members have the opportunity to participate in highly discounted genetic sequencing ($125 per sample) of the Kombucha SCOBY and broth. Fermentation scientists at Oregon State University and creating a database of microbial populations from commercial kombucha. Not only will you have the opportunity to see what your SCOBY is comprised of, but you will also be able to see your powerhouse compares to the average SCOBY. This is just the first step in understanding how the microbial population relates to kombucha features.
For more information in registering, visit: https://kombuchabrewers.org/kbi-osu-scoby-genomics-analyte-study/
Referenced Papers
- Coton, Monika, et al. “Unraveling Microbial Ecology of Industrial-Scale Kombucha Fermentations by Metabarcoding and Culture-Based Methods.” FEMS Microbiology Ecology, vol. 93, no. 5, 2017, pp. FEMS Microbiology Ecology, 2017, Vol. 93(5).
- Chakravorty, Somnath, et al. “Kombucha tea fermentation: Microbial and biochemical dynamics.” International journal of food microbiology 220 (2016): 63-72.
- El-Salam, S. S. A. (2012). 16S rRNA gene sequence detection of acetic acid bacteria isolated from tea kombucha. New York Science Journal, 5(3), 55-61.
- Jankovic, I., Stojanovic, M., 1994. Microbial and chemical composition, growth, therapeutical and antimicrobial characteristics of tea fungus. Mikrobiologija 33, 25 – 34.
- Liu, C-H., et al. “The isolation and identification of microbes from a fermented tea beverage, Haipao, and their interactions during Haipao fermentation.” Food Microbiology 13.6 (1996): 407-415.
- Marsh, Alan J., et al. “Sequence-based analysis of the bacterial and fungal compositions of multiple kombucha (tea fungus) samples.” Food microbiology 38 (2014): 171-178.
- Mayser, P., Gromme, S., Leitzmann, C., Gru¨nder, K., 1995. The yeast spectrum of the ‘tea fungus Kombucha’. Mycoses 38, 289 – 295
- Shade, Ashley, D. H. Buckley, and S. H. Zinder. “The kombucha biofilm: a model system for microbial ecology.” Final report on research conducted during the Microbial Diversity course. Marine Biological Laboratories, Woods Hole, MA (2011).
- Teoh, Ai Leng, Gillian Heard, and Julian Cox. “Yeast ecology of Kombucha fermentation.” International journal of food microbiology 95.2 (2004): 119-126.